BIO 349L, Project 2: Fetal Alcohol Spectrum Disorder
Abstract
Fetal Alcohol Spectrum Disorder (FASD) describes the range of phenotypic defects resulting from ethanol exposure during embryonic development. FASD is a product of complex interactions between genetic predispositions and the environment, often resulting in a spectrum of altered neurological and facial defects. Here we examined the effects exposure to 1% ethanol solution had on the development of facial bones in two mutant strains of zebrafish (131.5 and Au15). Specifically, we examined the length of the palatoquadrate (pq), ceratohyal (ch), and Mekel’s (m) bones, as well as the widths of the arches that they form. Our results found that these bones saw a statistically significant increase in bone length if the Au15 mutation was present, or if embryos were exposed to ethanol. These findings indicate an interaction between the environment and genetic mutations in the craniofacial development of zebrafish embryos.
Method
Embryos and treatment
Embryos were provided by the Eberhart lab, and all embryos were raised and cared for using established IACUC protocols approved by the University of Texas at Austin. Both mutant strains of zebrafish embryos (131.5 and Au15) were treated with 1% EtOH at 24 hpf, and embryos were allowed to develop for 4 days at 28.5°C. Embryos were then fixed in 2% PFA in PBS, and washed 1 x 10 min with 100 mM Tris pH 7.5 / 10 mM MgCl2.
Staining
Embryos were stained for cartilage (using Alcian blue stain) for one day, and consequently destained with a series of 80%, 50%, and 25% EthOH / 100 mM Tris pH 7.5 / 10 mM MgCl2 washes (3 x 5 min). Afterwards, embryos were bleached with 3% H2O2 / 0.5% KOH for 1 x 10 min and washed for 2 x 10 min with 25% glycerol / 0.1% KOH. Finally, embryos were stained for bone (using Alcian red stain) for 30 min, destained with 50% glycerol / 0.1% KOH, and imaged on stereoscopes.
Results
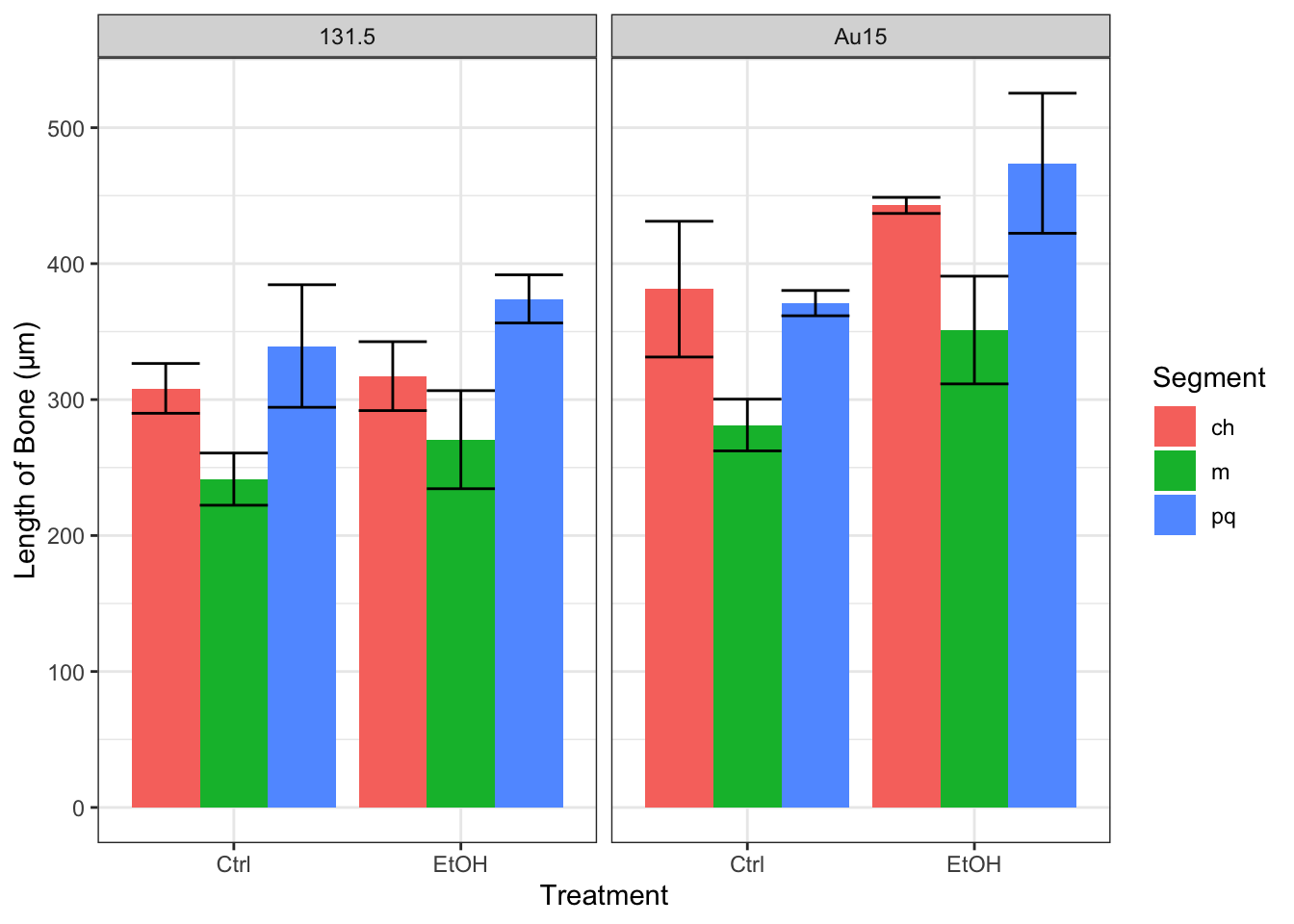
Our primary interest in this investigation was to explore the effects that ethanol exposure and genetic mutations had on the length of facial bones. We utilized the image processing software ImageJ to calculate a few key metrics from the images of our embryos. Among these metrics, we measured the length of three important facial bones (see Figure 1): Mekel’s (m; dark green), palatoquadrate (pq; light green), and ceratohyal (ch; red). Additionally, we measured the width that these bones spanned laterally (illustrated by the dotted/dashed lines in Figure 1).
Figure 1
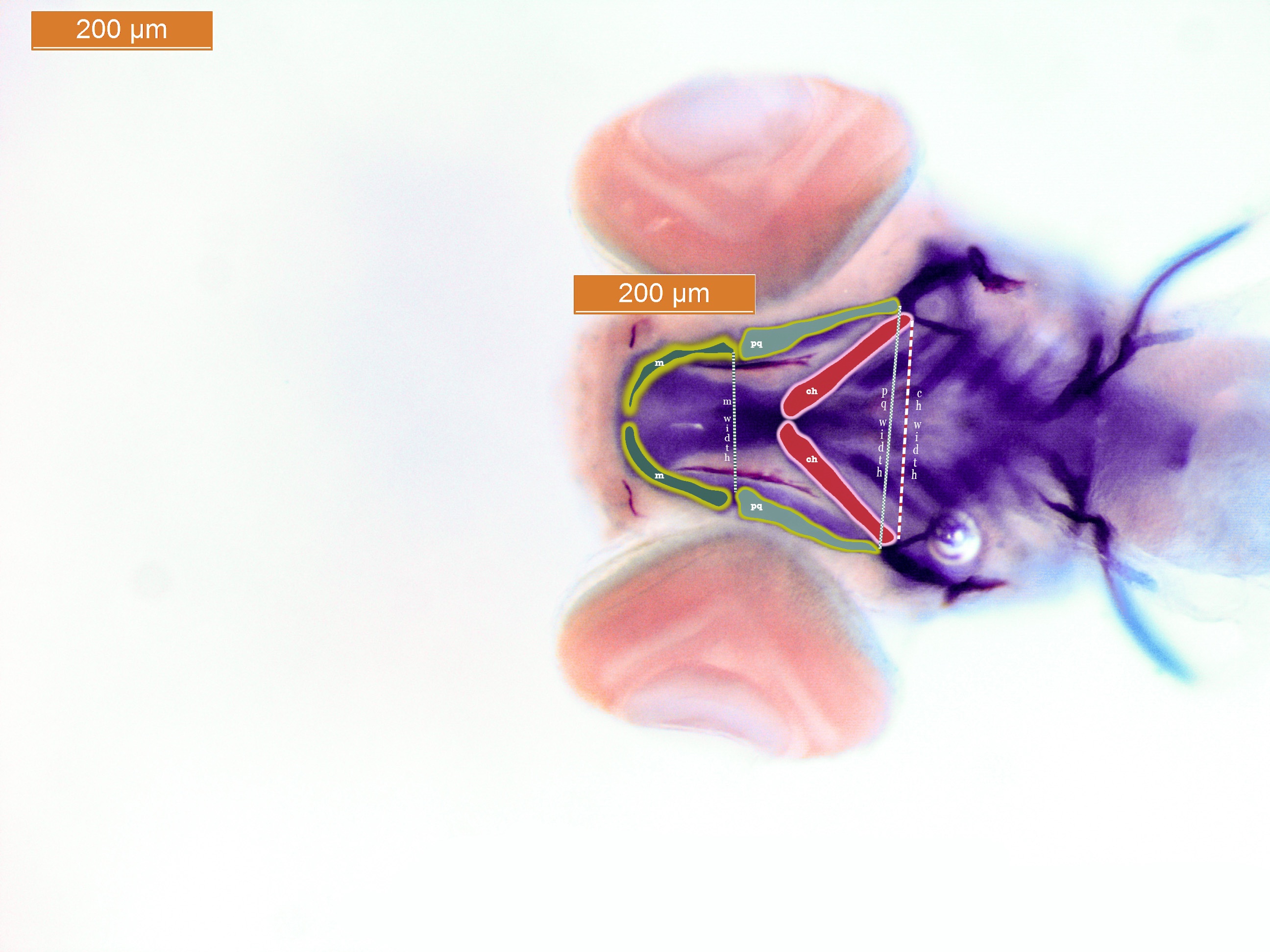
Figure 1
Linear Regression of Bone Length
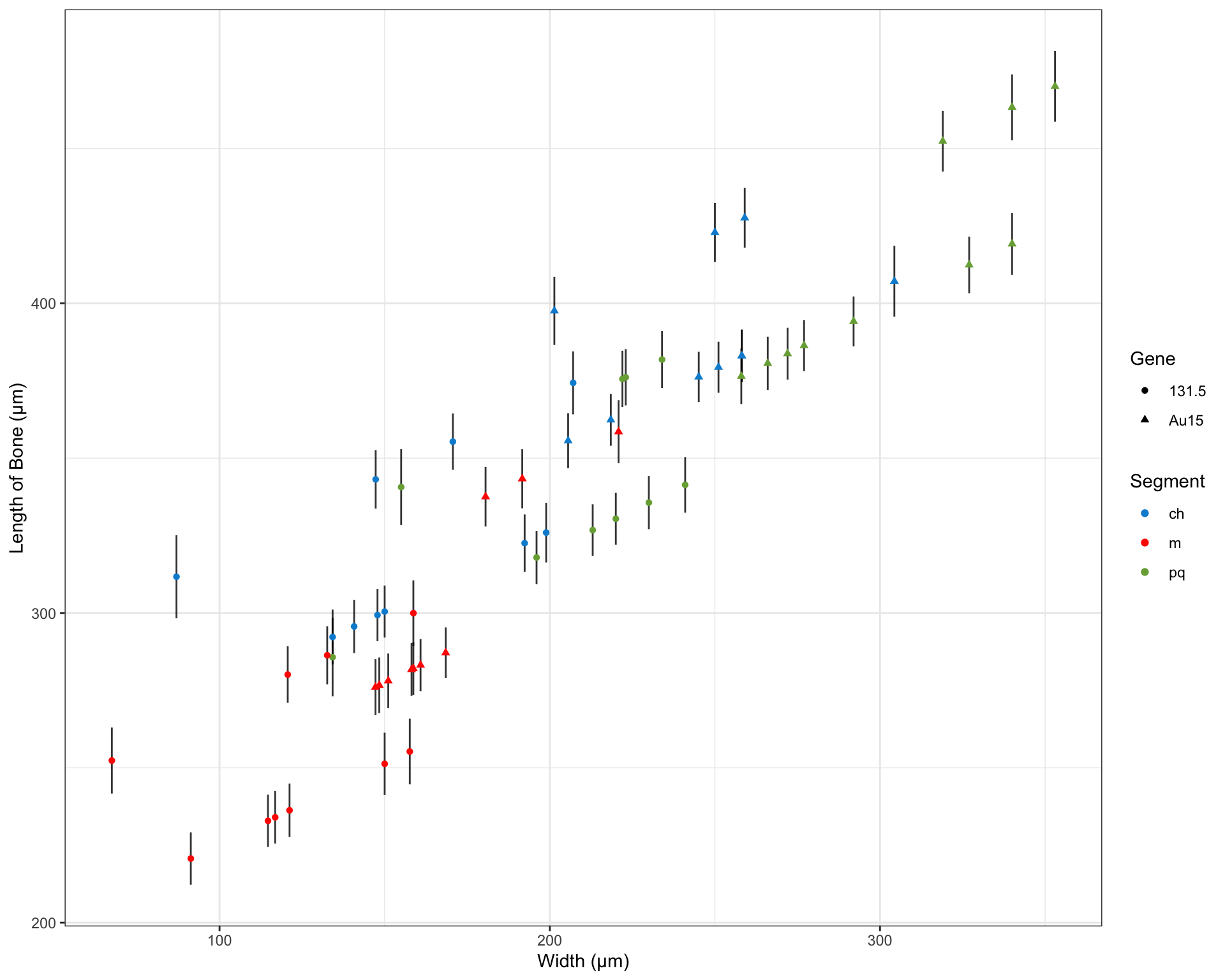
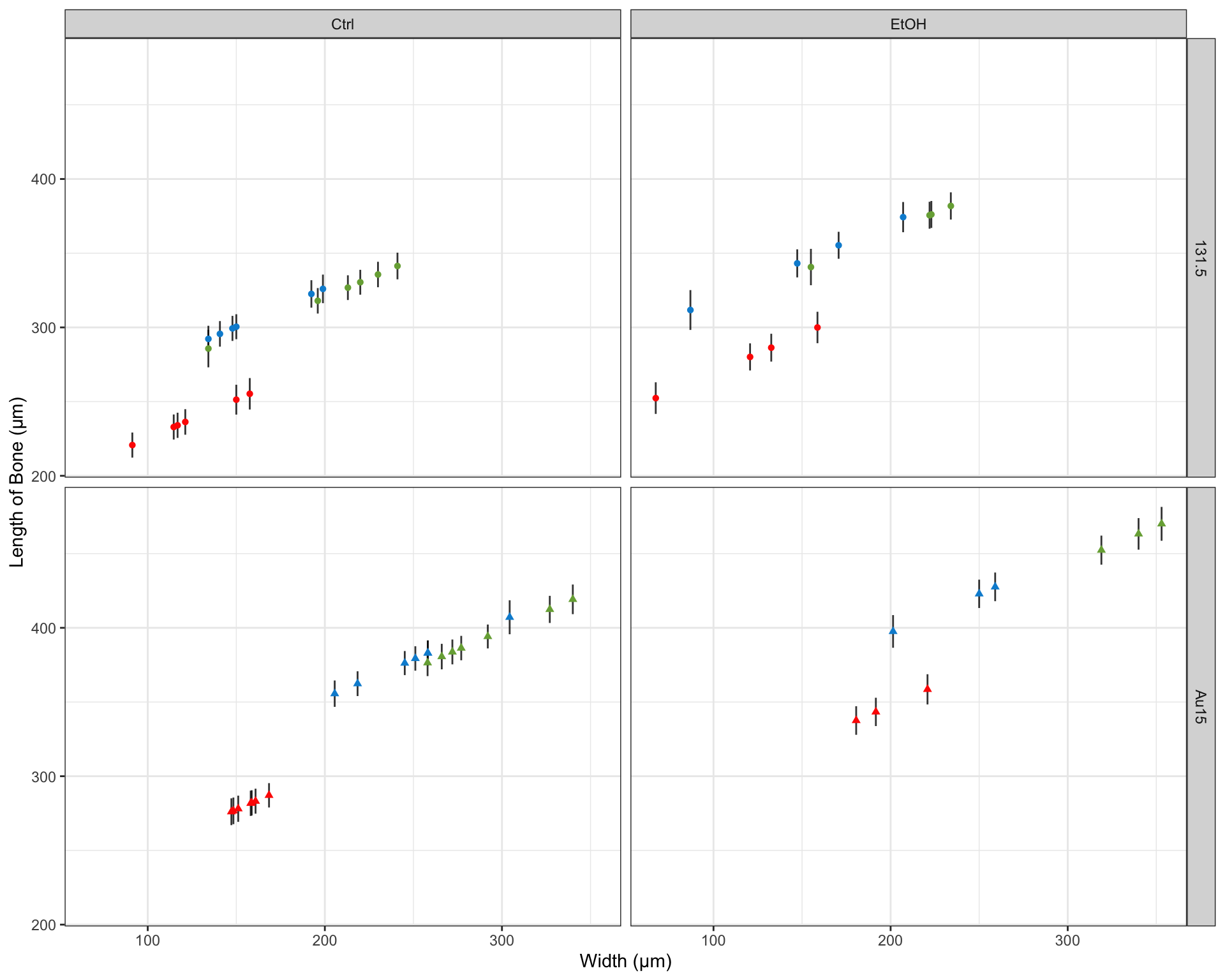
After performing a series of linear regressions modeling the length of the key facial bones described above, we found that treatment with ethanol, the width that the bones spanned (see Figure 1), and the mutations (131.5 or Au15) were all significant predictors of the bone length. Our model exhibited a residual standard error of 29.74 μm on 54 degrees of freedom (p-value: < 2.2 x 1016) and accounted for 80.3% of overall variance (R2=0.8197; adjusted 2=0.803). The summary of our model has been included below:
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.8197119 | 0.8030185 | 29.74064 | 49.10411 | 0 | 6 | -285.5264 | 585.0527 | 599.7132 | 47763.32 | 54 |
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 222.3 | 20.88 | 10.65 | 7.134e-15 |
| Width | 0.5212 | 0.1205 | 4.326 | 6.597e-05 |
| Segmentm | -49.16 | 11.54 | -4.26 | 8.243e-05 |
| Segmentpq | -6.513 | 11.45 | -0.5687 | 0.5719 |
| TreatmentEtOH | 44.11 | 8.185 | 5.389 | 1.592e-06 |
| GeneAu15 | 26.21 | 12.15 | 2.157 | 0.03549 |
df %>% group_by(Gene, Segment, Treatment) %>% summarise(m = mean(Length), sd = sd(Length)) %>%
ggplot(aes(x=Treatment, fill=Segment)) +
geom_bar(aes(y=m), stat="identity", position = "dodge") + geom_errorbar(aes(ymax=m+sd, ymin=m-sd), position = "dodge") +
scale_color_fivethirtyeight() +
labs(y="Length of Bone (µm)", x="Treatment") +
theme_bw() + theme(axis.title=element_text()) + facet_wrap("Gene")