BIO 349L, Project 3: Effect of Fluoride on Sea Urchin Development
Abstract
Fluoride has been linked to neurodevelopmental problems in human and animal models. Fluoride has previously been shown to inhibit acetylcholinesterases (AChE) in humans, and AChE expression in sea urchin models has been thought to mark the beginning of neuronal differentiation. Our aim was to explore the effects of early fluoride exposure on skeletal development, and determine if fluoride caused any changes in AChE expression/localization. To achieve this, we treated Lytechinus variegatus larvae with 4 mM sodium fluoride (NaF) for 48 h, and performed in situ hybridization for expression of the AChE mRNA. We found that exposure to NaF inhibited skeletal formation, decreased expression of AChE, and showed imprecise expression localization of AChE when compared to untreated controls. We concluded that NaF might interfere with the differentiation of mesenchymal cells, and consequently disrupt signaling from the ectoderm to the endoderm.
Introduction
In recent years, the adverse effects of high fluoride concentrations in drinking water has warranted increased scrutiny. Fluoride has been linked to neurodevelopmental problems in humans and animal models, although the mechanisms of action are still uncertain (Choi et al., 2012). Fluoride has been thought to inhibit some molecular functions involved with the nervous system. In particular, fluoride has been shown to inhibit acetylcholinesterases (AChE) in humans at levels as low as 15-25 μM (Cimasoni, 1966).
Sea urchins serve as a useful model organism for neurotoxicity, and fluoride has been known to inhibit skeleton formation and arm development. Young (1957) concluded that exposure to sodium fluoride (NaF) induced slight skeletal inhibition at 2 mM, total inhibition at 5 mM, and no inhibition at 1 mM. Larval skeleton formation and arm development is thought to be dictated, in part, by mesenchymal cells attached to the skeleton tip. These cells are localized during the early stages of development, and are critical messengers between the ectoderm and the skeleton (Gustafson & Wolpert, 1961).
Incidentally, AChE expression rapidly increases during gastrulation, and expression has been shown to be localized in mesenchymal cells (Ozaki, 1974). This increase in AChE expression is believed to be the beginnings of neuronal differentiation in the ectoderm (Akasaka et al., 1986) and might play a role in signaling for skeletal development.
The aim of this study was to explore the effects of NaF exposure on AChE expression in L. variegatus larvae. Using data measuring arm formation under different concentrations of NaF and intervals of exposure (Vokes, 2015), we found that exposure to NaF before gastrulation results in major skeleton deformations. Exposure after/during gastrulation results in far fewer deformations, indicating that NaF induces its effects during a key window in early development.
To identify genes modulated by NaF, we utilized RNA-Seq in L. variegatus to measure mRNA expression in the presence of NaF, and electronically annotated select genes that may help explain how NaF inhibit arm formation. Among these annotated genes was Lv-AcheL_3, which encodes for AChE. Finally, to investigate the localization of AChE expression, we performed in situ hybridization of Lv-AcheL_3 in larvae that were exposed to 4 mM of NaF for 24 h.
We found that NaF exposure both decreased expression of AChE, as well as inhibited skeletal formation. In untreated larvae, AChE expression was localized around the mesenchyme cells associates with the spine, oral lobe, and postoral arms. In NaF treated larvae, however, AChE expression was not as clearly localized, and appeared to be disrupted.
Methods
RNA-Seq and Electronic Annotation
To investigate the effect of NaF on mRNA expression, RNA-Seq was performed on larvae in the presence and absence of NaF. Expression levels were then compared, preserving only reads with a p-value < 0.05. Reads were then mapped to the Lytechinus variegatus genome with a 20% alignment.
After discarding outliers, 289 genes were successfully mapped to the
L. variegatus genome. Subsequently, the top 50 genes (by largest
Log2(Fold Change) magnitude) were electronically annotated, yielding their
predicted protein families, Gene Ontology (GO) biological process and GO
molecular function.
Fertilization, NaF Treatment, and Fixation
Gametes of L. variegatus (provided by KP Aquatics) were obtained using KCl injection and fertilized. Control larvae were then cultured in artificial sea water (ASW), and treated larvae were cultured in NaF (4 mM) + ASW. Both larvae conditions were cultured for 24 h at room temperature, following which they were fixed for 1 h with 4% PFA in ASW at room temperature. Finally, larvae were washed with MetOH, and stored at 4°C until in situ hybridization.
In Situ Hybridization with Lv-AcheL_3 probe
Fixed larvae were rehydrated with PBST and digested with 4ug/ml Proteinase K in PBST @ RT for 15 min. Following the digest, larvae were washed with PBST 2x3’, re-fixed with 4% PFA for 10 min, and washed again with PBST.
Larvae were incubated at 46°C in Hyb solution for 1 h, after which the Hyb solution was replaced with a probe for our gene of interest (Lv-AcheL_3) in hyb solution and incubated overnight at 46°C. The following day, the probe was removed, washed with SSC with Tween (SSCT) for 1 h @ 37°C, and rinsed with PBST.
Larvae were then blocked for 1 h with blocking solution, and stored in dilute block solution with alkaline phosphatase labeled anti-DIG antibody at @ 4°C overnight. The following day, larvae were washed with PBST and washed in staining buffer 3x5’. Finally, larvae were incubated in NBT/BCIP stain @ RT for 30 min in the dark, and the stained larvae were imaged with DIC and polarized light microscopy.
Results
Skeletal development is disrupted by NaF exposure
Our aim was to discover if disruptions caused to skeletal development was concentration or time specific. We accomplished this by analysing the conditional distribution of the number of arms that developed under differing concentrations of NaF and exposure intervals (Vokes, 2015).
We found that exposure to NaF at 4 mM and 2 mM resulted in a much smaller proportion of larvae that developed both arms (32.1% and 34.9%, respectively; Fig 1a) when compared to unexposed controls (79.4% developed both arms; Fig 1a). However, we did find that the timing of NaF exposure was important: Larvae exposed before 24 h had a smaller proportion develop normal skeletons (20%) compared to those exposed after 24 h (61.4%) or unexposed controls (79.4%) [Fig 1b].
Notably, we only see a marginal decrease in normal skeletal development when the concentration of NaF is doubled (2 mM → 4 mM ≈ ∆ -2.8%) or when the exposure interval is doubled beyond the first 24 h (∆ - 1.7%). From this data, we concluded that skeletal development was most disrupted when larvae were exposed to at least 2 mM NaF before gastrulation (~ 24 h).
NaF decreases expression for nucleosome assembly genes
After performing RNA-seq for NaF exposed larvae, we electronically annotate select genes to determine how NaF might affect biological processes. Among our 52 annotated genes, 24 genes returned no predictions for the GO biological process. While we only annotated a fraction of the RNA-seq data, a few clear patterns emerged.
Among our 289 mRNAs (with a p-value < 0.05), 207 mRNAs saw decreased expression
when exposed to NaF. This can be best visualized by the grey histograms in
Fig 3abc, or by the disproportionate number of points below the solid line in
Fig 2b.
The biological process GO predictions (Figure 3a) from many of these downregulated mRNAs indicate that nucleosome assembly might be negatively influenced by NaF exposure. In fact, among the annotated mRNAs with a biological process prediction, 54% matched to this GO term. Furthermore, IPR Scan annotations indicate a number of these mRNAs are members of the histone family of proteins (Figure 3b) and GO predictions for their molecular functions indicate they could be involved with DNA binding and have protein heterodimerization activity (Figure 3c).
Acetylcholinesterase expression is reduced in NaF exposed larvae
In order to explore the relationship between mRNA expression levels and observed
skeletal development in larvae exposed to NaF, we performed in situ
hybridization of a selected gene from the RNA-seq data. Our gene of interest,
Lv-AcheL_3 (acetylcholinesterase-like-3), was severely inhibited in the presence
of NaF (Log2FC = -1.927 at p < 5e-5).
As expected, overall expression was lower in NaF treated larvae (Figure 4d) than in untreated controls (Figure 4b). We also observed a much higher incidence of skeletal deformations in NaF treated larvae (Figures 4c, 4d) than in untreated (Figures 4a, 4b), falling in agreement with our previous analysis of the Vokes data (2015).
Moreover, Lv-AcheL_3 expression in untreated larvae appeared to be localized in the areas we expect to find mesenchyme cells (Figure 4b), while NaF treated larvae had much less distinctive patterning (Figure 4d).
Discussion
In this study, we found that exposure to NaF at levels greater than 2 mM severely impairs skeletal development. The analysis of the Vokes (2015) data and observations from the in situ hybridization clearly indicate that exposed larvae are less likely to develop both arms normally. This falls in line with previous research indicating that NaF exposure caused slight skeletal inhibition (Young, 1957).
From the Vokes data, we also concluded that NaF exposure occurring before gastrulation produces the most severe skeletal inhibition (Fig 1b). Incidentally, AChE expression rapidly increases during gastrulation, and expression has been shown to be localized in mesenchymal cells associated with the spine, oral lobe, and postoral arms (Akasaka et al., 1986; Ozaki, 1974). Furthermore, our RNA-Seq data indicates that Lv-AcheL_3 expression is decreased in the presence of NaF (Fig 2), and our in situ hybridization further supports this observation (Figure 4d).
Gustafson & Wolpert (1961) found that mesenchymal cells that eventually become the endoderm dictate larval arm development, and supporting research has indicated that NaF decreases expression of AChE in mesenchymal cells (Ozaki, 1974). It is therefore possible that in the early stages of development, NaF interferes with the differentiation of mesenchymal cells that would become neurons (Akasaka et al., 1986). This, in turn, could disrupts the ectoderm (e.g. the nervous system) ability to differentiate properly, and consequently disrupt skeletal formation by the endoderm.
Limitations and Future Research
A significant limitation of this study is the narrow scope of genes selected. We only investigated one of the many genes that saw a change in expression with NaF, and it’s unlikely that AChE expression is the only factor at play with disrupted skeletal development. For this reason, we cannot determine causation by this relationship. While it is possible that NaF disrupts AChE expression directly (thereby inhibiting mesenchymal cells differentiation and skeletal development), this seems unlikely. NaF likely inhibits some other cellular process in the early stages, which in turn inhibits Lv-AcheL_3 expression in mesenchymal cells and skeletal development.
Author: Hunter Ratliff @HunterRatliff1
Figures
Figure 3
Fig 3a
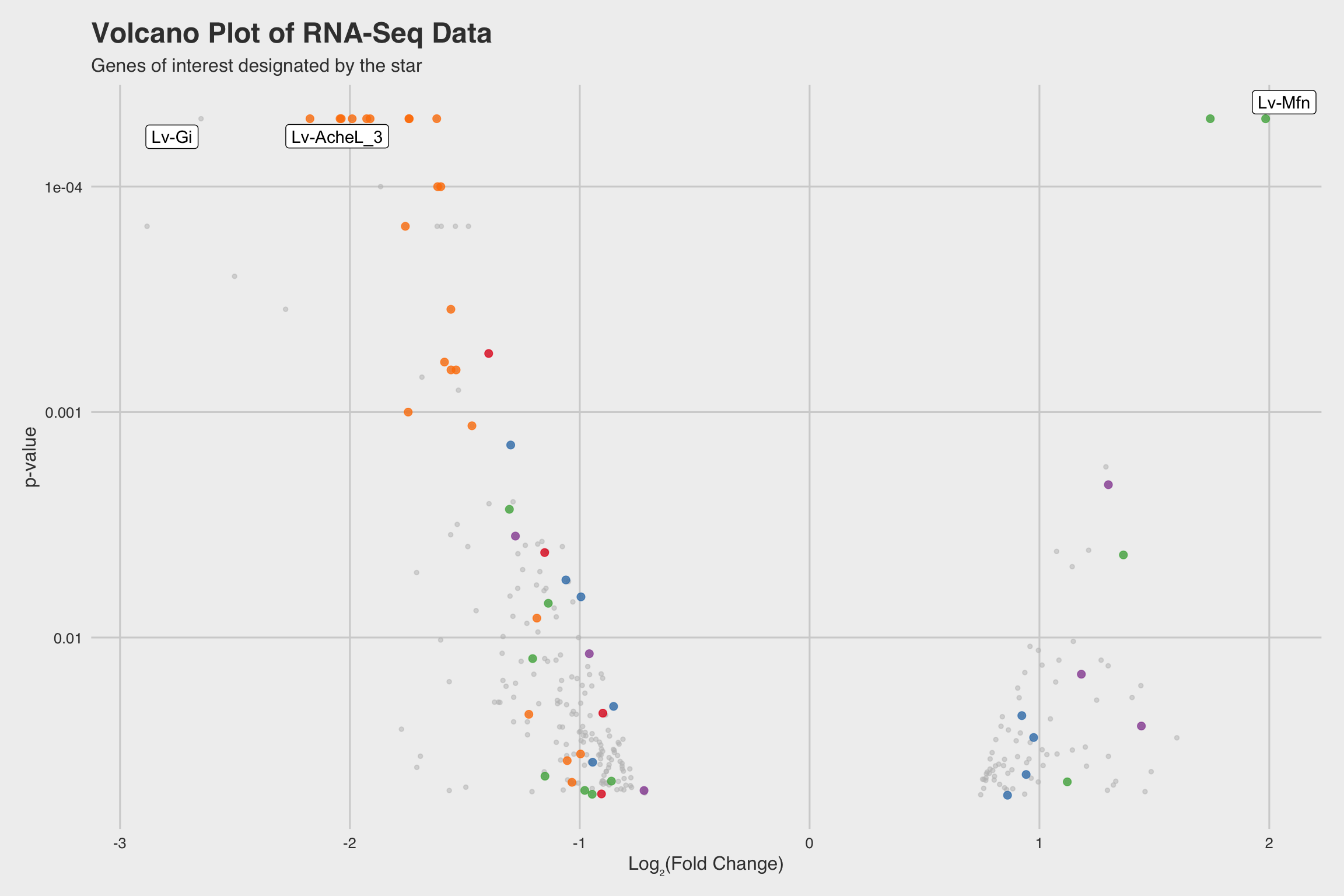
Figure 1: Fig 3a
Fig 3b
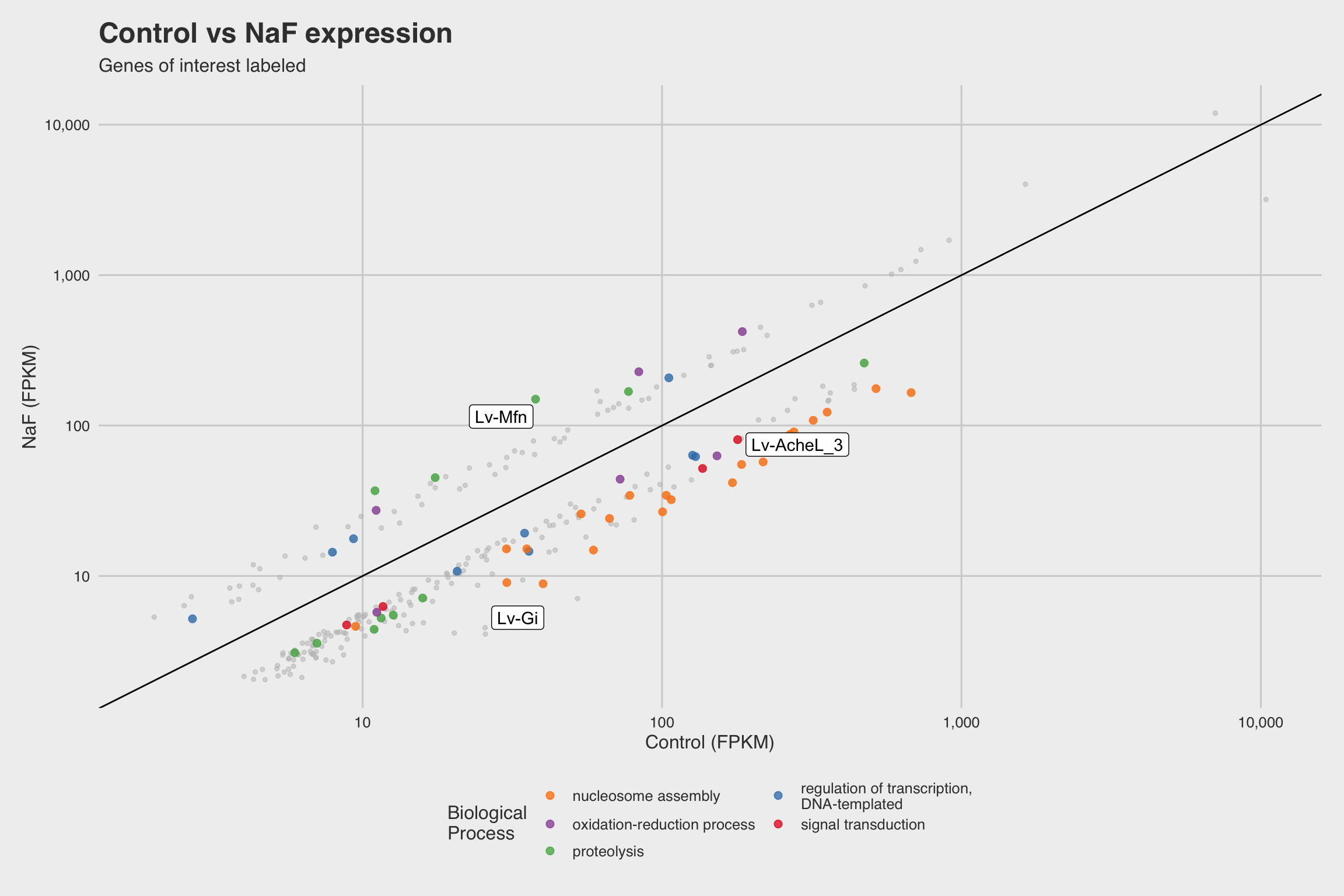
Figure 2: Fig 3b
Fig 3c
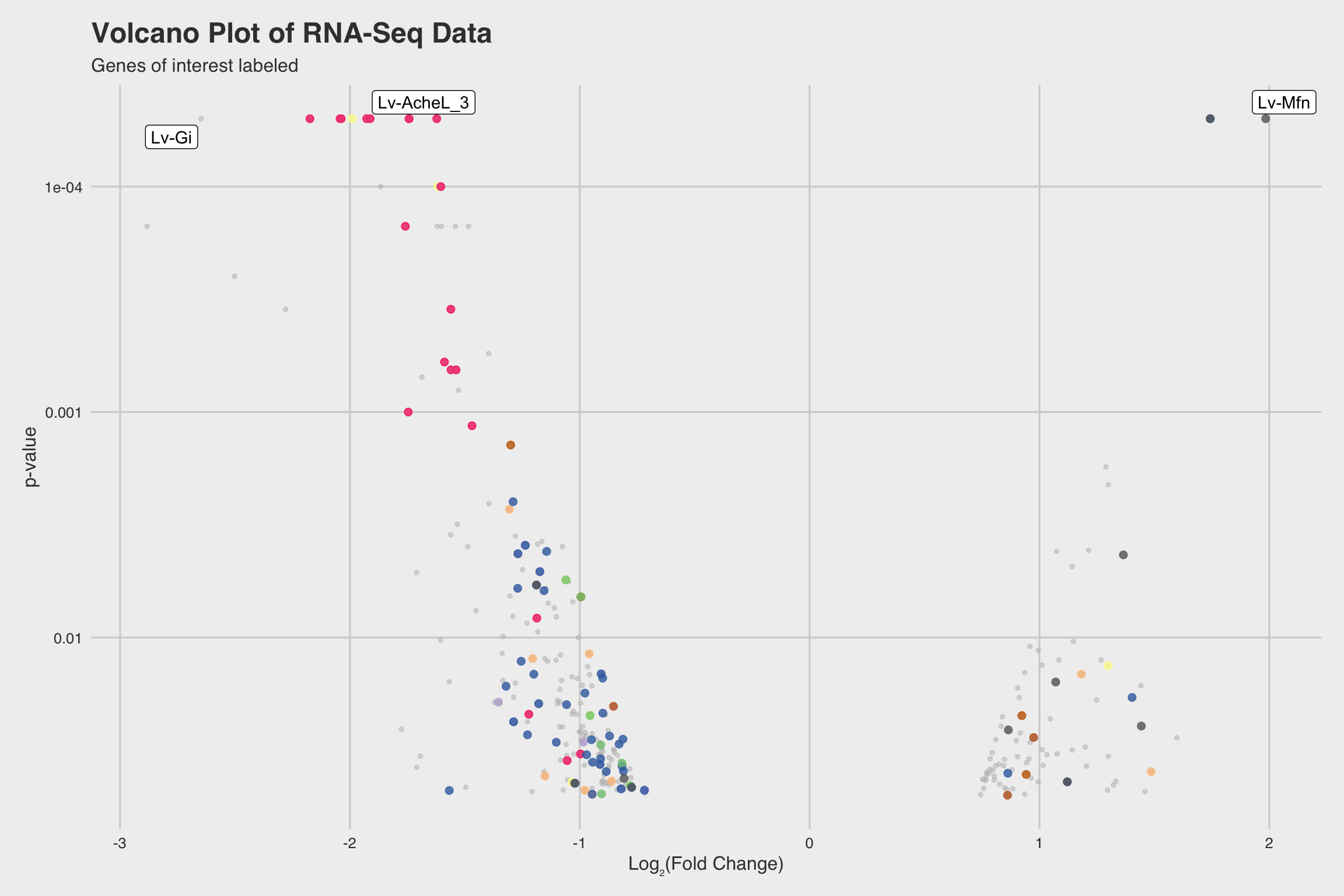
Figure 3: Fig 3c
Fig 3d
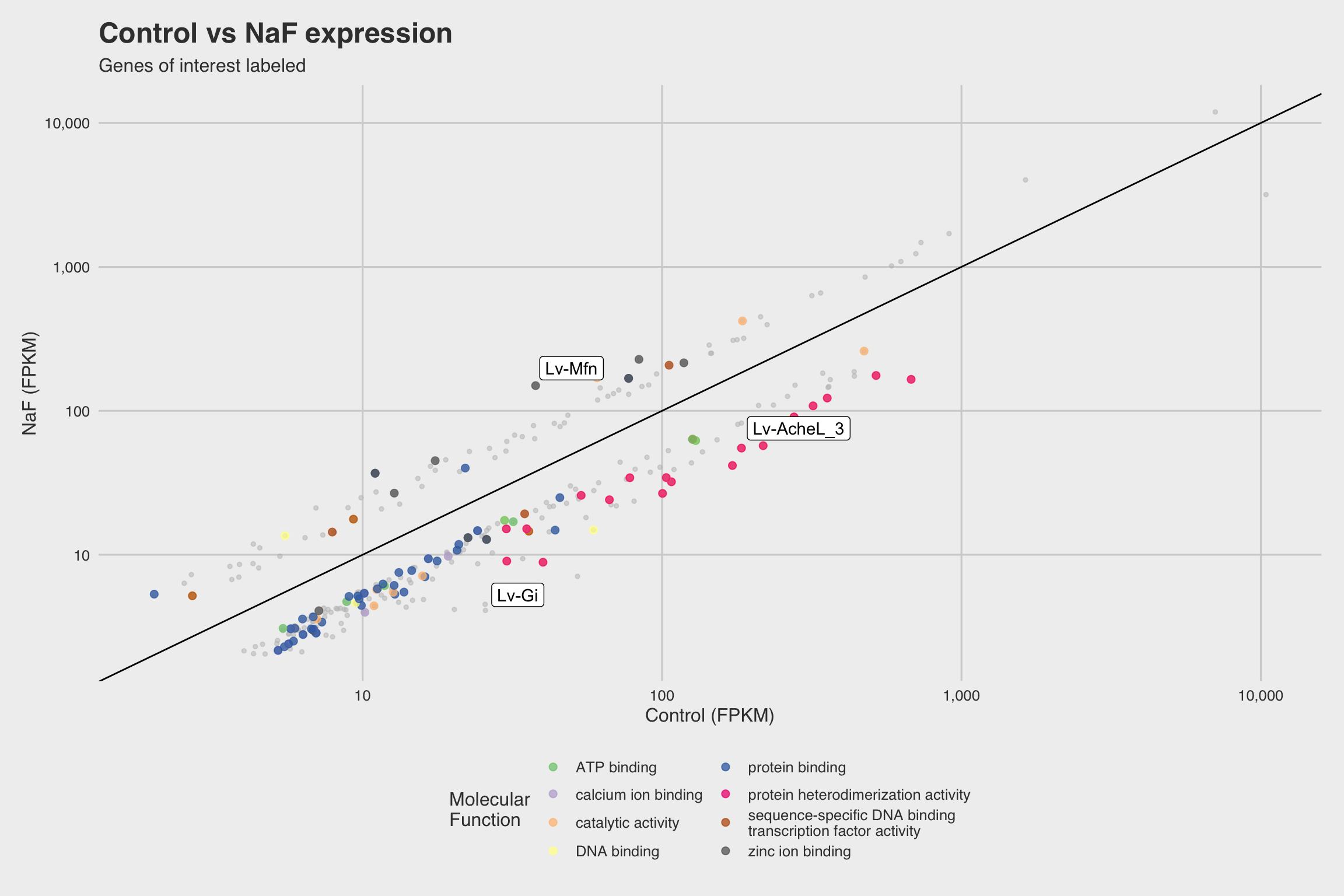
Figure 4: Fig 3d
Fig 3e
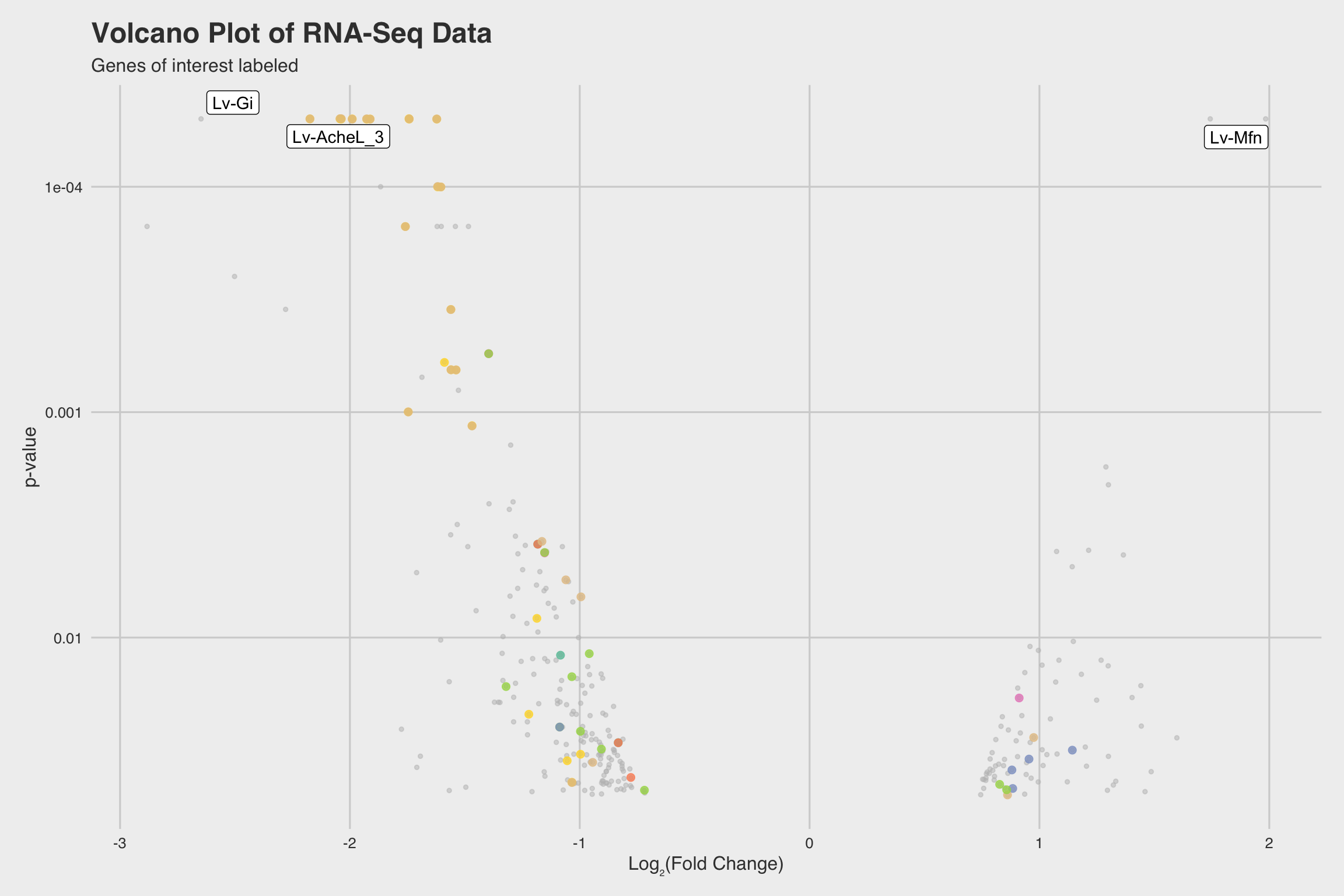
Figure 5: Fig 3e
Fig 3f
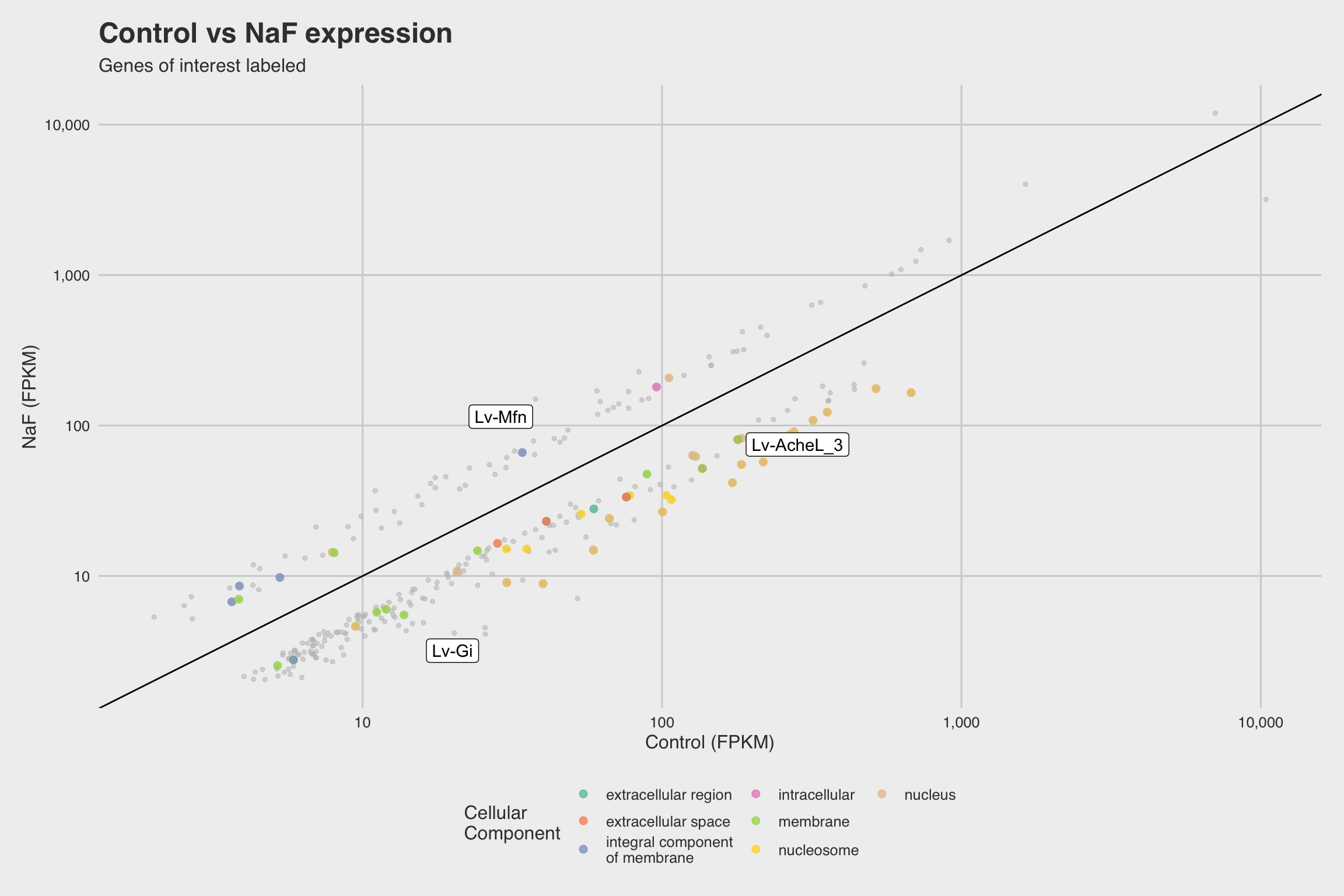
Figure 6: Fig 3f
Figure 4
Fig 4a
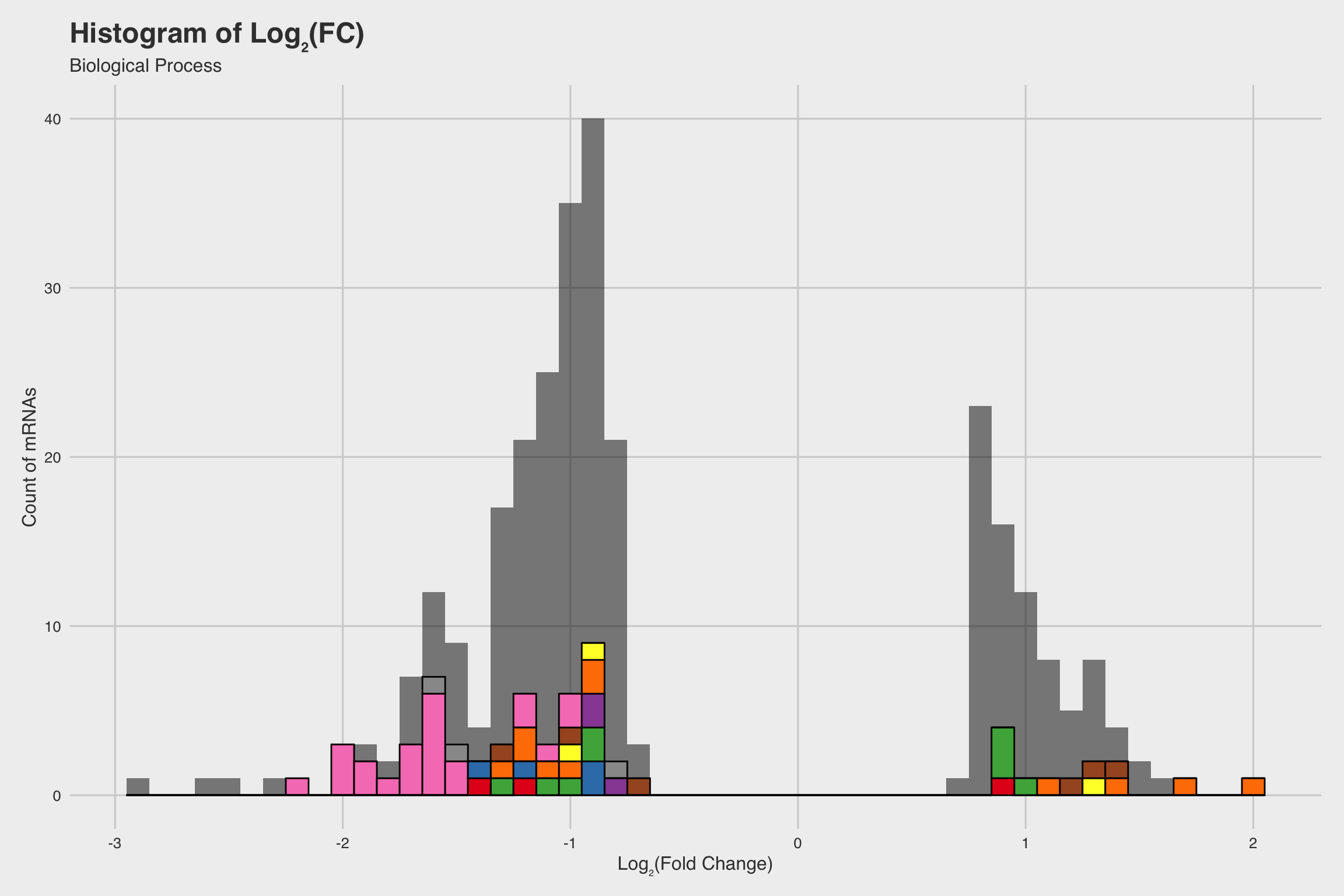
Figure 7: Fig 4a
Fig 4b
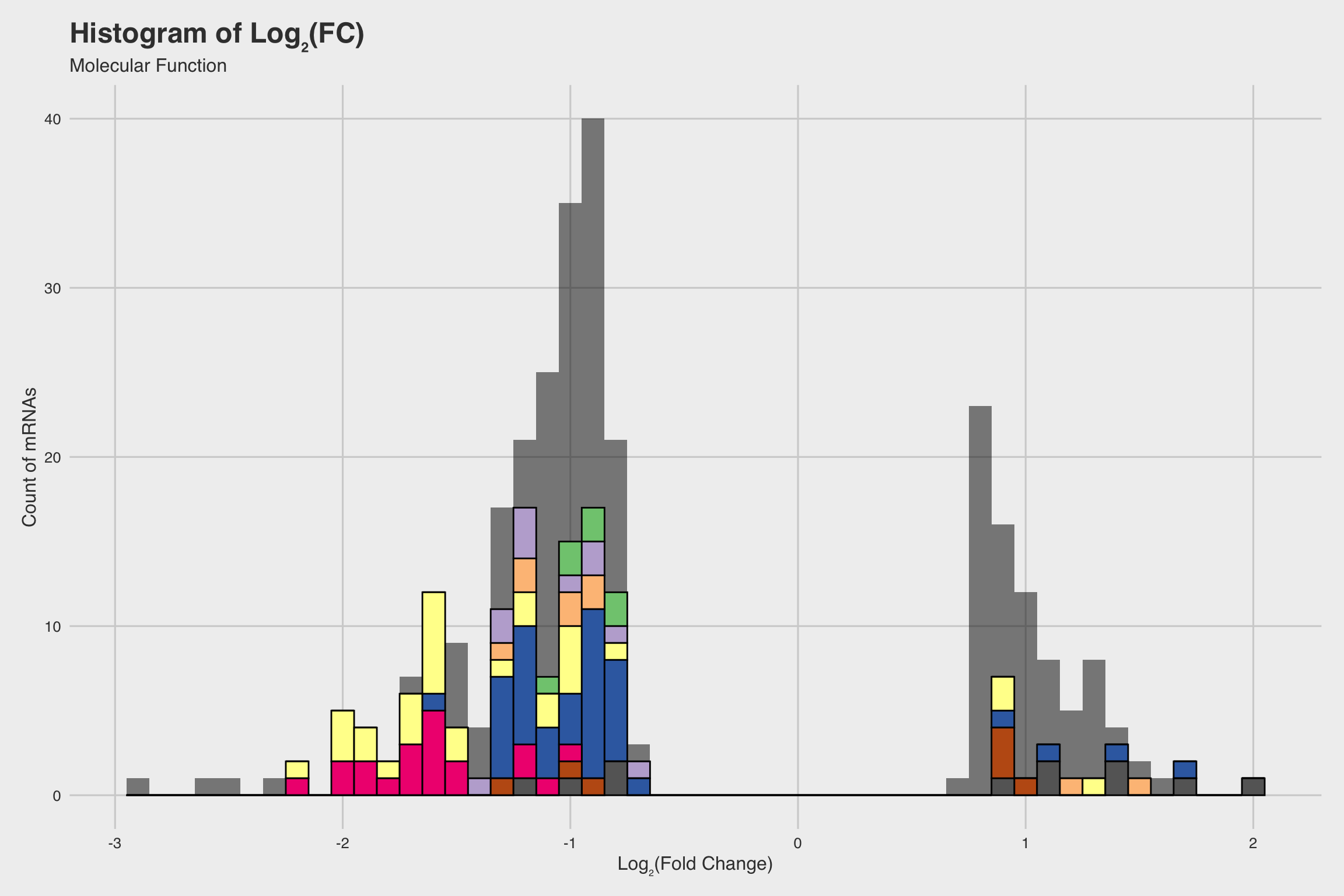
Figure 8: Fig 4b
Fig 4c
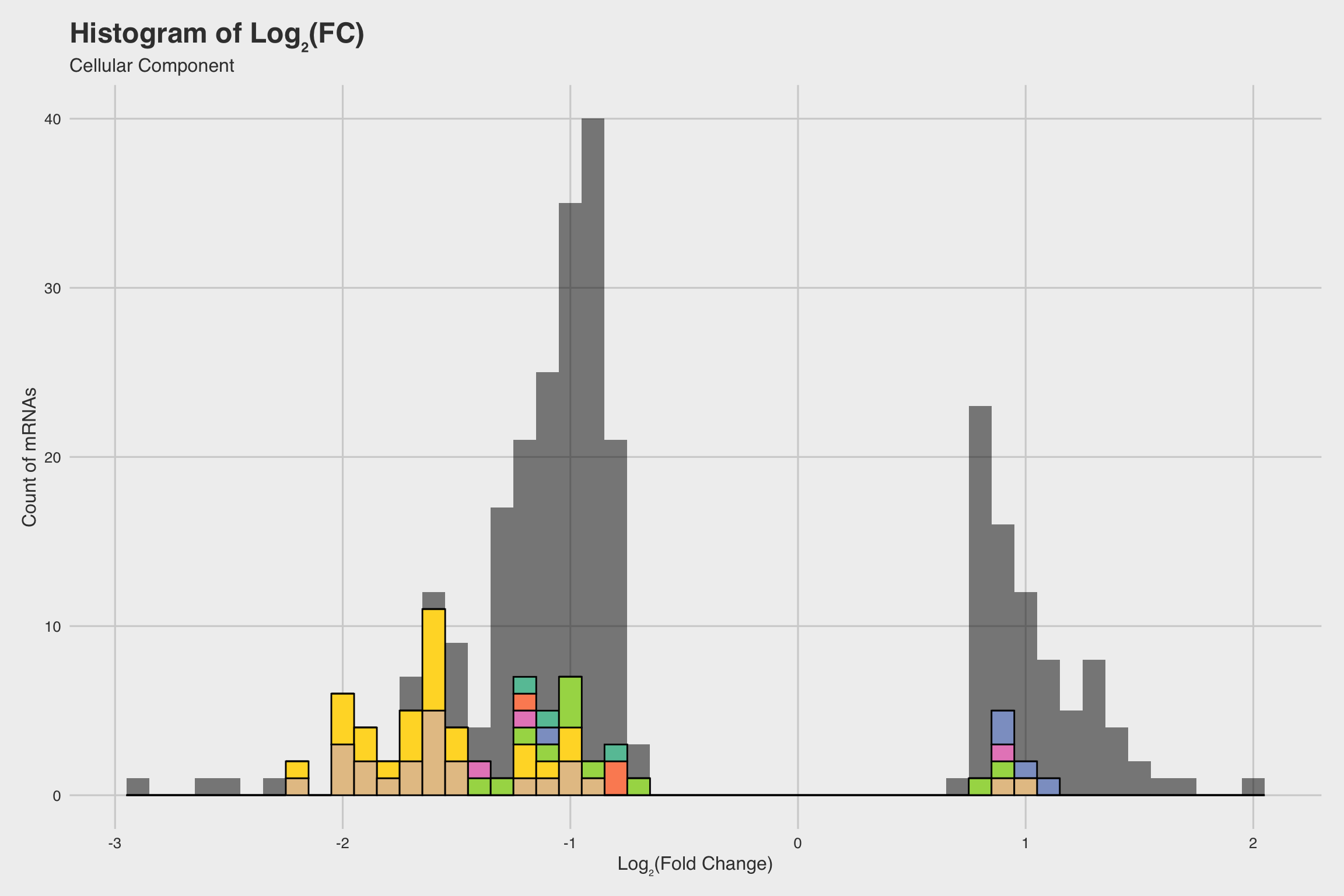
Figure 9: Fig 4c
Fig 4d
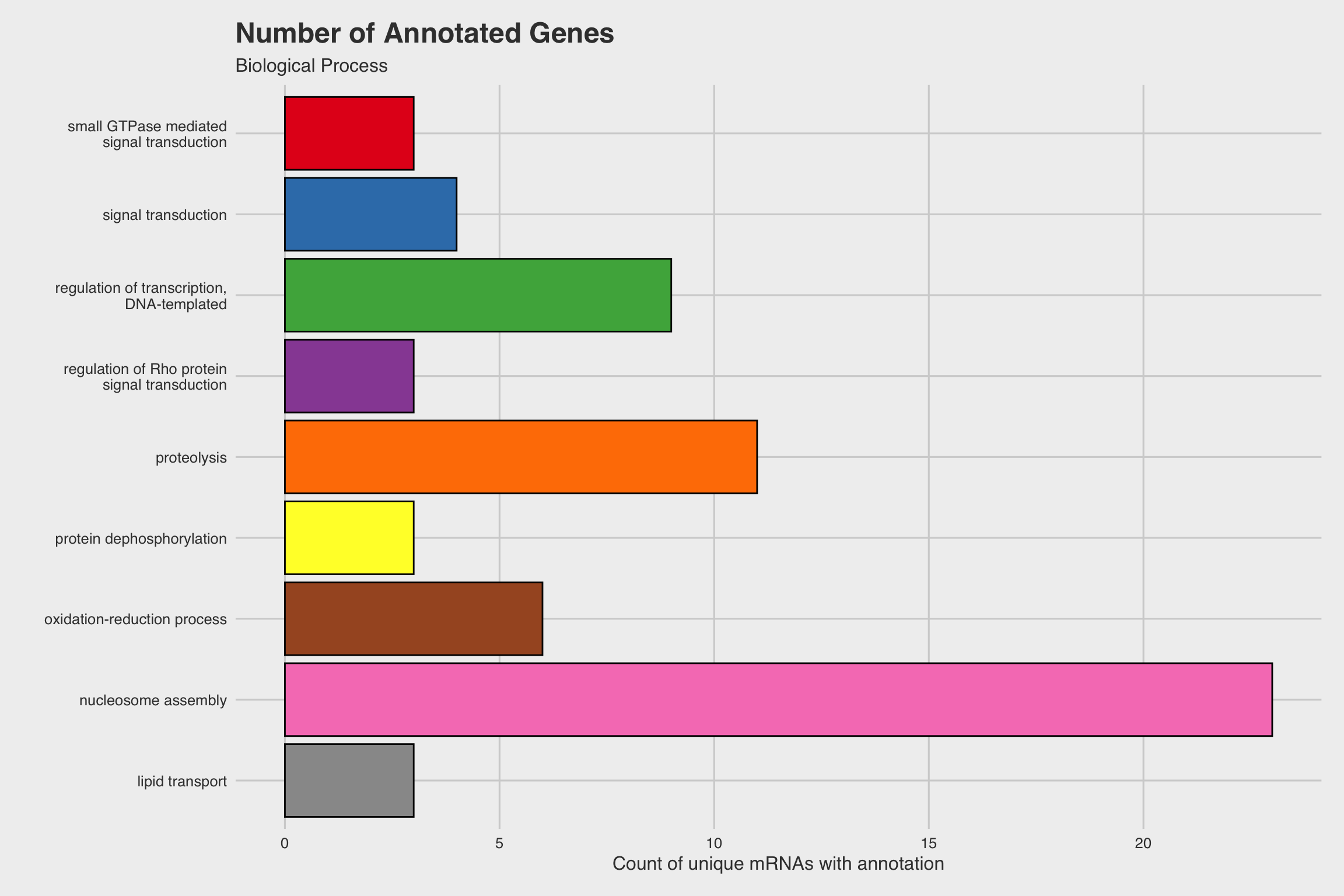
Figure 10: Fig 4d
Fig 4e
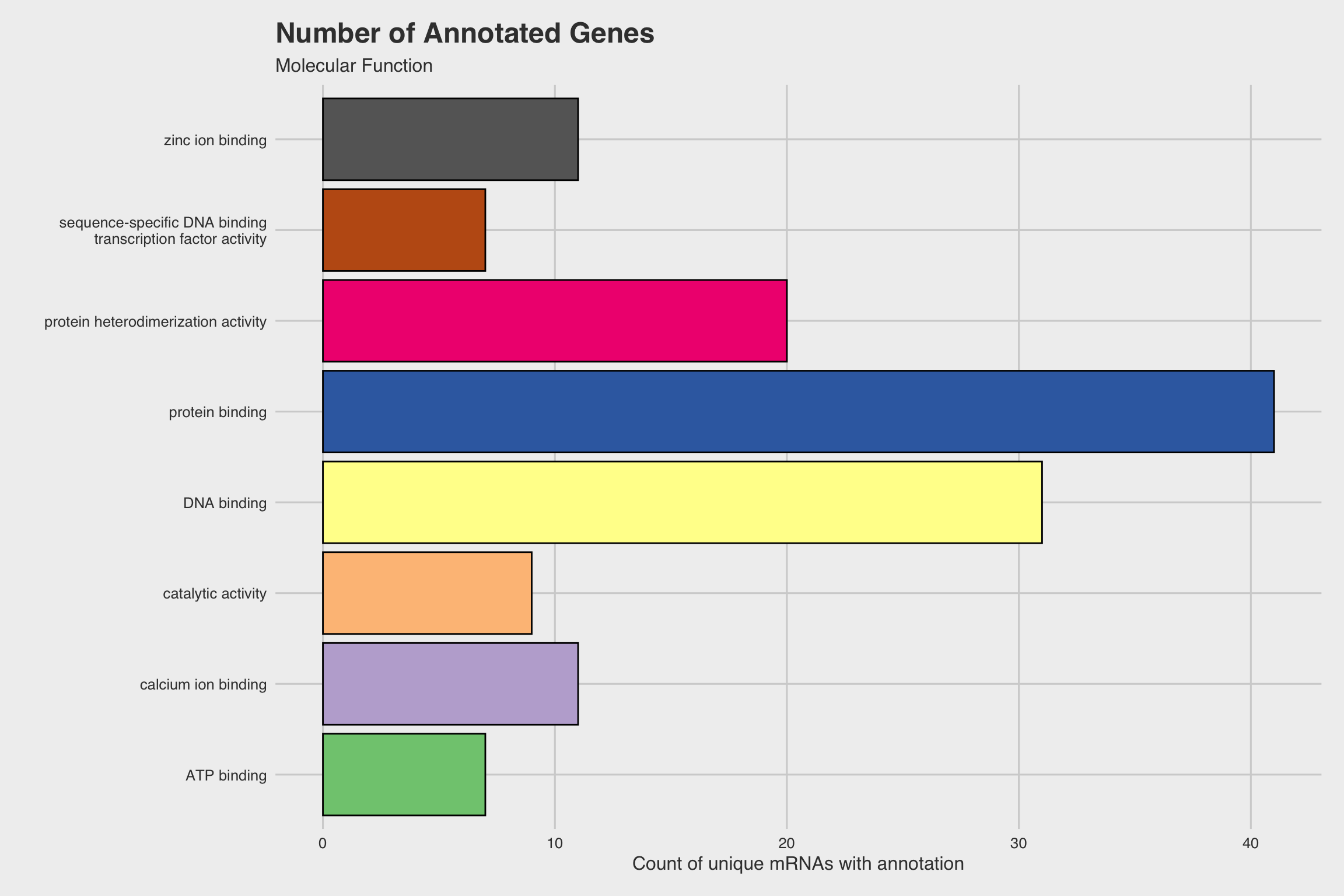
Figure 11: Fig 4e
Fig 4f
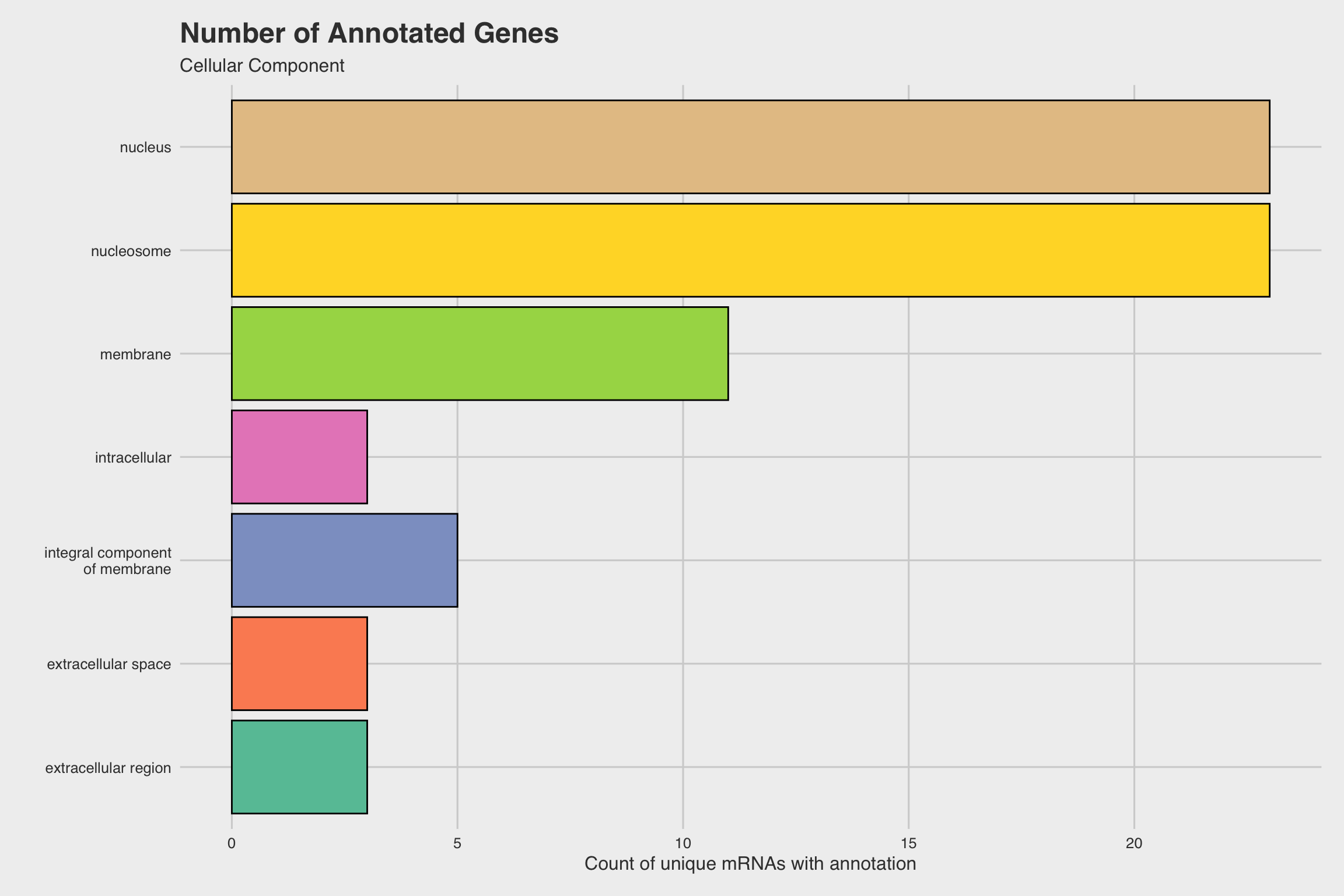
Figure 12: Fig 4f
Fig 4g
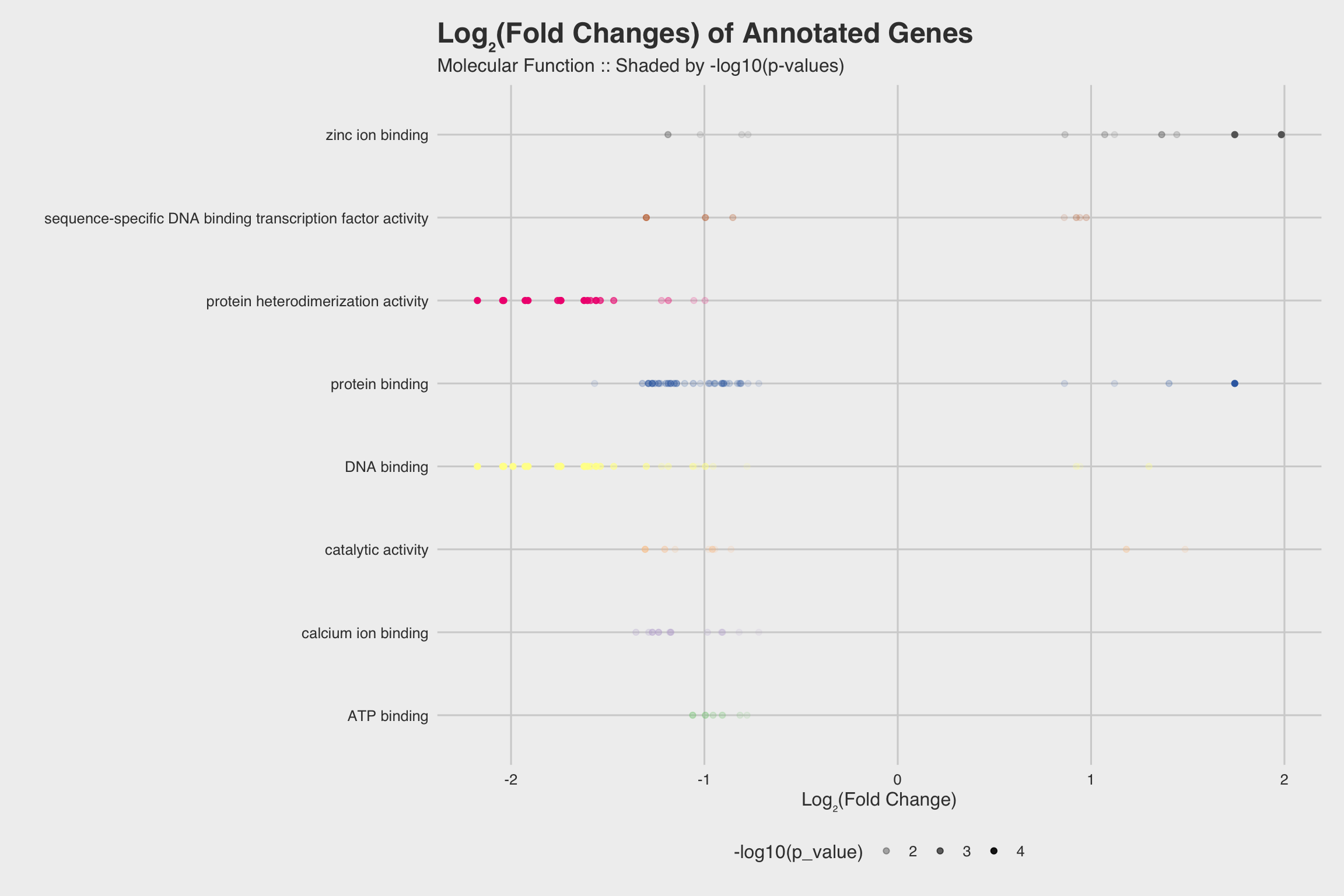
Figure 13: Fig 4g
Fig 4h
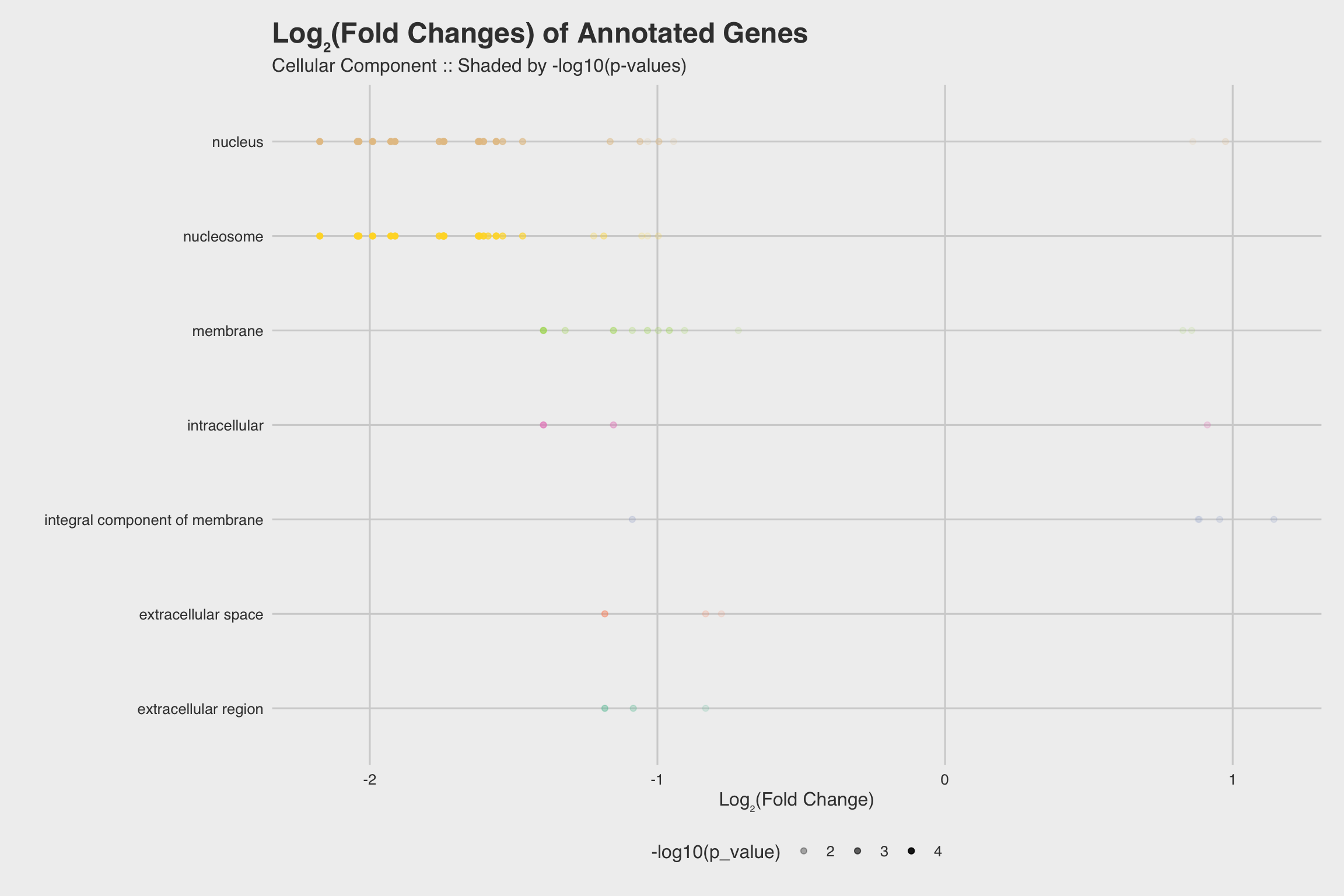
Figure 14: Fig 4h
License
--- LICENSE ---
Copyright (C) 2016 Hunter Ratliff
This program is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
You should have received a copy of the GNU General Public License
along with this program. If not, see <http://www.gnu.org/licenses/>.This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see http://rmarkdown.rstudio.com.
In the spirit of Reproducible Research, below is the information About the R Session at the time it was compiled1:
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 3.6.3 (2020-02-29)
## os macOS Catalina 10.15.4
## system x86_64, darwin15.6.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/Chicago
## date 2020-06-12
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.0)
## backports 1.1.6 2020-04-05 [1] CRAN (R 3.6.2)
## blogdown 0.18 2020-03-04 [1] CRAN (R 3.6.0)
## bookdown 0.18 2020-03-05 [1] CRAN (R 3.6.0)
## callr 3.4.3 2020-03-28 [1] CRAN (R 3.6.2)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.0)
## cli 2.0.2 2020-02-28 [1] CRAN (R 3.6.0)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.0)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.0)
## curl 4.3 2019-12-02 [1] CRAN (R 3.6.0)
## data.table 1.12.8 2019-12-09 [1] CRAN (R 3.6.0)
## desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.0)
## devtools * 2.2.2 2020-02-17 [1] CRAN (R 3.6.0)
## digest 0.6.25 2020-02-23 [1] CRAN (R 3.6.0)
## dplyr * 0.8.5 2020-03-07 [1] CRAN (R 3.6.0)
## ellipsis 0.3.0 2019-09-20 [1] CRAN (R 3.6.0)
## evaluate 0.14 2019-05-28 [1] CRAN (R 3.6.0)
## fansi 0.4.1 2020-01-08 [1] CRAN (R 3.6.0)
## farver 2.0.3 2020-01-16 [1] CRAN (R 3.6.0)
## forcats 0.5.0 2020-03-01 [1] CRAN (R 3.6.0)
## foreign 0.8-75 2020-01-20 [1] CRAN (R 3.6.3)
## fs 1.4.1 2020-04-04 [1] CRAN (R 3.6.2)
## ggplot2 * 3.3.0 2020-03-05 [1] CRAN (R 3.6.0)
## ggrepel * 0.8.2 2020-03-08 [1] CRAN (R 3.6.0)
## ggthemes * 4.2.0 2019-05-13 [1] CRAN (R 3.6.0)
## glue 1.4.0 2020-04-03 [1] CRAN (R 3.6.2)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.0)
## haven 2.2.0 2019-11-08 [1] CRAN (R 3.6.0)
## highr 0.8 2019-03-20 [1] CRAN (R 3.6.0)
## hms 0.5.3 2020-01-08 [1] CRAN (R 3.6.0)
## htmltools 0.4.0 2019-10-04 [1] CRAN (R 3.6.0)
## knitr 1.28 2020-02-06 [1] CRAN (R 3.6.0)
## labeling 0.3 2014-08-23 [1] CRAN (R 3.6.0)
## lifecycle 0.2.0 2020-03-06 [1] CRAN (R 3.6.0)
## magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.0)
## memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.0)
## openxlsx 4.1.4 2019-12-06 [1] CRAN (R 3.6.0)
## pillar 1.4.3 2019-12-20 [1] CRAN (R 3.6.0)
## pkgbuild 1.0.6 2019-10-09 [1] CRAN (R 3.6.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 3.6.0)
## pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.6.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 3.6.0)
## processx 3.4.2 2020-02-09 [1] CRAN (R 3.6.0)
## ps 1.3.2 2020-02-13 [1] CRAN (R 3.6.0)
## purrr 0.3.4 2020-04-17 [1] CRAN (R 3.6.2)
## R6 2.4.1 2019-11-12 [1] CRAN (R 3.6.0)
## RColorBrewer 1.1-2 2014-12-07 [1] CRAN (R 3.6.0)
## Rcpp 1.0.4.6 2020-04-09 [1] CRAN (R 3.6.3)
## readr 1.3.1 2018-12-21 [1] CRAN (R 3.6.0)
## readxl 1.3.1 2019-03-13 [1] CRAN (R 3.6.0)
## remotes 2.1.1 2020-02-15 [1] CRAN (R 3.6.0)
## rio * 0.5.16 2018-11-26 [1] CRAN (R 3.6.0)
## rlang 0.4.6 2020-05-02 [1] CRAN (R 3.6.2)
## rmarkdown 2.1 2020-01-20 [1] CRAN (R 3.6.0)
## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.0)
## scales 1.1.0 2019-11-18 [1] CRAN (R 3.6.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.6.0)
## stringi 1.4.6 2020-02-17 [1] CRAN (R 3.6.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 3.6.0)
## testthat 2.3.2 2020-03-02 [1] CRAN (R 3.6.0)
## tibble 3.0.0 2020-03-30 [1] CRAN (R 3.6.2)
## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 3.6.2)
## usethis * 1.5.1 2019-07-04 [1] CRAN (R 3.6.0)
## vctrs 0.3.0 2020-05-11 [1] CRAN (R 3.6.2)
## withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.0)
## xfun 0.13 2020-04-13 [1] CRAN (R 3.6.3)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 3.6.0)
## zip 2.0.4 2019-09-01 [1] CRAN (R 3.6.0)
##
## [1] /Library/Frameworks/R.framework/Versions/3.6/Resources/libraryThe date compiled may differ from the date the code was written because it get’s re-run when it’s uploaded to the site↩